Forum Replies Created
-
AuthorPosts
-
I agree with Y Soun.
Besides the ease and convenience for the patient, Liquid biopsy can detect cancer very early before the appearance of any signs and before any imaging technique can do. This is of utmost importance when we are concerned with monitoring the efficacy of treatment and testing for relapses.
Circular RNA (circRNA) is a type of single-stranded RNA which, unlike linear RNA, forms a covalently closed continuous loop, with the 3′ and 5′ ends normally present in an RNA molecule have been joined together. Because circular RNA does not have 5′ or 3′ ends, it is resistant to exonuclease-mediated degradation and is more stable than linear RNA.
Circular RNAs are highly represented in the eukaryotic transcriptome. Thousands of endogenous circRNAs have been discovered in mammalian cells. They are generated from exonic or intronic sequences of precursor mRNA.
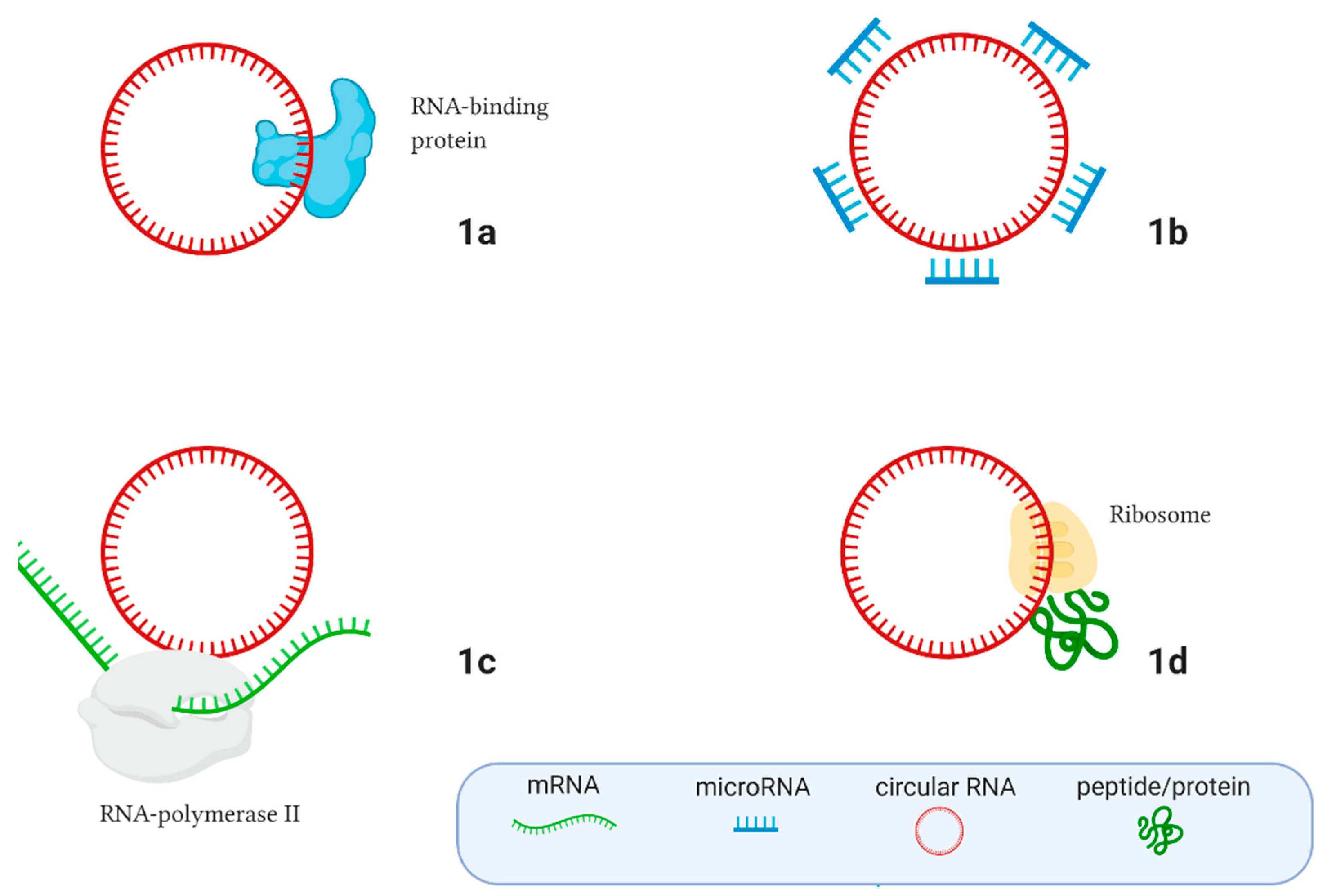
Circular RNAs have been shown to play important roles as microRNA sponges, regulators of gene splicing and transcription, RNA-binding protein sponges, and protein/peptide translators. They function in various human diseases, particularly cancers, and may function as predictive biomarkers and therapeutic targets for cancer treatment.
Reference: Tang, X., Ren, H., Guo, M., Qian, J., Yang, Y., & Gu, C. (2021). Review on circular RNAs and new insights into their roles in cancer. Computational and structural biotechnology journal, 19, 910–928. https://doi.org/10.1016/j.csbj.2021.01.018
Metabolome is the sum of all metabolites (small molecules). The suffix “ome” means the sum of all. Hence, as stated by John Brown above, the genome is the sum of all genetic material (all DNA), transcriptome is the sum of all transcripts (all RNA), and proteome is the sum of all proteins.
And yes, the metabolome differs from one tissue to another, and from one condition to another. It can form a fingerprint for a specific tissue under specific conditions.
The suffix “omics” means the science of studying the specific set. Therefore, metabolomics is the science of studying tissue metabolites under different conditions. It is important for studying tissue biology, and it has various useful applications. For example, it helps in studying mechanisms of disease development and devising treatment strategies.
For more on “omics”, you can visit the Wikipedia site.
The Glossary of this site gives a brief description of some of these terms.
CRISPR-Cas9 is originally a defense mechanism of bacteria against invading phages. It is now used as a gene editing technique, which was recognized by the Nobel Prize in Chemistry in 2020.
CRISPR is an acronym for Clustered Regularly Interspaced Short Palindromic Repeats. It is a family of DNA sequences found in the genomes of prokaryotic organisms. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e., anti-phage) defense system of prokaryotes and provide a form of acquired immunity.
Cas9 (or “CRISPR-associated protein 9”) is an endonuclease enzyme. It uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA that are complementary to the CRISPR sequence.
This defense mechanism forms the basis of a technology known as CRISPR-Cas9 that can be used to edit genes within organisms. Cas9 is used with a guide RNA to cut double stranded DNA at a specific site identified by the guide RNA. By manipulating the nucleotide sequence of the guide RNA, the artificial Cas9 system could be programmed to target any DNA sequence for cleavage.
This editing process has a wide variety of applications including basic biological research, development of biotechnological products, and diagnosis and treatment of diseases.
Go to this site News to see that CRISPR-Cas9 technology has been used for treatment of sickle cell disease and targeting cancer cells for destruction.
To read more about the applications of CRISPR-Cas9, click here.
-
AuthorPosts
