Researchers at the University of Cambridge, UK reported a general approach, Chem-map, to establish drug-chromatin binding maps. These are in situ interaction maps for small molecules that bind to cellular genomic DNA or chromatin-associated proteins. They illustrated the method with three distinct interaction modalities for molecules that either target a chromatin protein, a DNA secondary structure or intercalate DNA.
Importance of mapping the drug-chromatin binding
Small molecules that interact with cellular DNA formed the basis for early anticancer molecules that became widely deployed in the clinic. A greater understanding of genome structure and function has created new opportunities for intervening with disease states by targeting cellular DNA or the associated proteins with small molecules. It is essential to validate target engagement for molecular probes and therapeutic drugs. Characterizing drug-target engagement is essential to understand how small molecules influence cellular functions. Mapping where a small molecule binds chromatin can help explain the downstream genetic or epigenetic response.
How Chem-map works for profiling drug-chromatin interaction
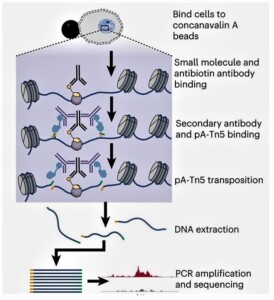
The researchers introduced this method, utilizing small-molecule-directed transposase Tn5 tagmentation. In permeabilized cells, a precomplex of biotinylated small molecules (yellow) and antibiotin antibody (black) bind to the chromatin target (protein or DNA). Then a secondary antibody (purple) tethering of pA–Tn5 transposomes (light blue) was recruited to the drug binding sites. Addition of Mg2+ activates the transposomes and integrates adapters (green and orange) in the proximity of the drug-binding sites. After DNA purification, genomic fragments with adapters at both ends are enriched via PCR, which allows the genome-wide identification of drug-binding sites by next-generation sequencing.
Implications of Chem-map profiling of drug-chromatin binding
Many approved, widely used drugs thought to act via a DNA targeting modality have, remarkably, not yet had their cellular molecular targets validated. In situ maps of small-molecule interactions with DNA and chromatin proteins in intact cells would provide valuable insights into the mode of action of this family of drugs and enhance our ability to exploit the genome as a therapeutic target. They provide insights that will enhance understanding of genome and chromatin function and therapeutic interventions.
Source:
Yu, Z., Spiegel, J., Melidis, L. et al. Chem-map profiles drug binding to chromatin in cells. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-022-01636-0
See also:
Bioinformatics Terms – Glossary – Bioinformatics Hub
Precision Therapy of Cancer: A New Platform – Bioinformatics Hub
MinION Nanopore Sequencer – Bioinformatics Hub
Go to the News Board