Now that the whole genome sequencing (WGS) is available, and genes can be edited, linking all observed health and disease traits to corresponding genes is a dream to chase. To understand how diversity in the human genome sequence influences phenotypic traits we need detailed analysis of genetic and phenotypic variation. Over the past decade, insights into this relationship have been obtained from large population studies. A recent article in Nature describes mass whole genome sequencing, associating identified variants with various human traits.
Mass whole genome sequencing for individuals from the UK Biobank
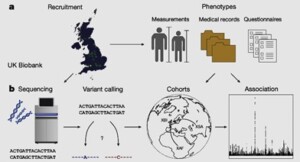
The researchers determined the whole-genome sequences of 150,119 individuals from the UK Biobank and identified more than 600 million sequence variants. The comprehensive data identify novel associations with human traits and show the functional importance of sequence variants inside and outside protein-coding regions.
The UK Biobank contains in-depth phenotypic information about 500,000 people from across the United Kingdom. Some previous studies of the biobank have focused on single-nucleotide polymorphisms (SNPs), but SNP arrays typically capture only a small fraction of common variants in the genome. Other studies have performed whole-exome sequencing (WES), which is limited to protein-coding regions and so reveals only a small proportion (2–3%) of sequence variation. The WES data also miss genetic variants outside coding exons, yet abundant evidence suggests that such variants can have important biological functions. Full genome sequences of members of the UK Biobank provides a unique opportunity to study how sequence diversity influences human diseases and other traits.
The UK population is diverse in its genetic ancestry and includes people born all around the globe. The researchers were able to define three cohorts within the UK Biobank based on genetic ancestry: a large British–Irish cohort and smaller African and South Asian cohorts. The African and South Asian cohorts each contain more than 9,000 people, and represent some of the largest available sets of whole-genome sequences of these ancestries. These data are likely to be valuable for identifying variants involved in disease and other traits.
The continued effort to obtain both WGS and rich phenotypic data for all 500,000 participants in the UK Biobank promises to vastly increase our understanding of the function of the non-coding genome. However, the biobank overwhelmingly contains data from individuals of British–Irish ancestry, so it is important that equally detailed WGS and analysis is carried out on diverse populations around the world.
See also: Telomere-to-Telomere (T2T): The first complete human genome
Go to the News Board
Pingback: Lung Cancer Genes Found - Bioinformatics Hub